In the last two decades, nearly 60 small molecule kinase inhibitors have been approved by the FDA for oncology indications. Although the human kinome consists of around 500 kinases, the inhibitors that are currently approved for cancer treatment act primarily through approximately 20 different kinases. Consequently, the majority of these kinases are targeted by multiple inhibitors. Identifying differences in the biochemical and cellular activities of comparator inhibitors can help in selecting the best suited inhibitor for therapy and identifying novel potential drug response biomarkers.
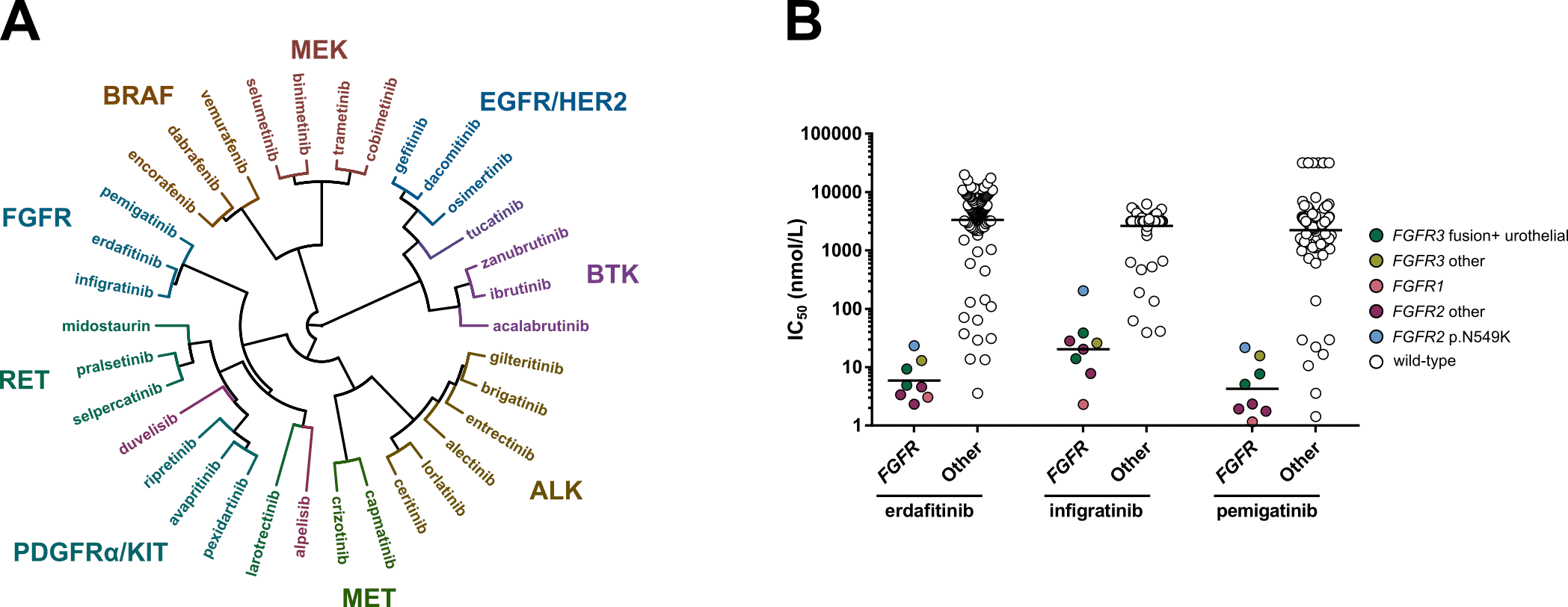
Figure. A) Hierarchical clustering based on the IC50 fingerprints of 21 recently approved kinase inhibitors and 13 previously approved drugs acting on the same targets, as determined in viability assays with 134 cancer cell lines. B) Scatterplots of the IC50 distribution of the three FGFR inhibitors across FGFR-altered and wild-type cell lines.
In our new study, which is published in Frontiers in Oncology, we profiled 21 newly FDA approved kinase inhibitors and 13 previously approved comparators on panels of 255 kinases and 134 cell lines (Figure A). This work follows up on our previous studies in which we profiled all kinase inhibitors that were approved for oncology indications until 2018 (Uitdehaag et al. (2014), Uitdehaag et al. (2019)). In our latest study we uncover important similarities and differences between inhibitors that act on the same primary target.
We found that comparing the cellular IC50 fingerprint of the kinase inhibitors clusters most inhibitors together which act on the same primary target. Interestingly, some inhibitors clustered with inhibitors that act on a different primary target, such as the FLT3 inhibitor gilteritinib and the TRK inhibitor entrectinib, which clustered with ALK inhibitors. By combining the biochemical and cellular profiling data of gilteritinib and entrectinib it was uncovered that, besides their primary targets, both inhibitors also potently inhibit the ALK kinase and ALK-altered cell lines.
The article also describes interesting observations for the three approved FGFR inhibitors. At time of writing of the publication, these FGFR inhibitors were only approved for either urothelial carcinoma harboring genomic alterations in FGFR2 and FGFR3 (erdafitinib), or FGFR2-altered cholangiocarcinoma (pemigatinib and infigratinib). We show that all three drugs potently target a wide range of FGFR-altered cell lines, irrespective of the specific underlying FGFR alteration or disease type (Figure B). Among these cell lines was an FGFR1-altered AML cell line. Interestingly, the approval of one of the FGFR inhibitors (pemigatinib) has been very recently expanded to also include this indication. These results illustrate that extensive in vitro profiling of kinase inhibitors can aid in identifying new patient populations that may benefit from treatment with these inhibitors.